 |
GeNN
2.2.3
GPU enhanced Neuronal Networks (GeNN)
|
 |
GeNN
2.2.3
GPU enhanced Neuronal Networks (GeNN)
|
In this tutorial we will learn to add synapsePopulations to connect neurons in neuron groups to each other with synatic models. As an example we will connect the ten Hodgkin-Huxley neurons from tutorial 1 in a ring of excitatory synapses.
First, copy the files from Tutorial 1 into a new directory and rename them to new names, e.g.
Now, we need to add a synapse group to the model that allows to connect neurons from the Pop1 group to connect to other neurons of this group. Open tenHHRingModel.cc, change the model name inside,
Now we need additional initial values and parameters for the synapse and post-synaptic models. We will use the standard NSYNAPSE weightupdate model and EXPDECAY post-synaptic model. They need intial variables and parameters as follows:
If an array is not needed we set it to the NULL pointer. Here there are for example no synaptic parameters and no initial values for the post-synaptic mechanism. We can then add a synapse population at the end of the modelDefinition(...) function,
The addSynapsePopulation parameters are
const char *name: The name of the synapse population, here "Pop1self" int sType: The type of synapse to be added, we here use the predefined typse NSYNAPSE. See Built-in Models for all available predefined synapse types. int sConn: The type of synaptic connectivity, here DENSE which means we will provide a full connectivity mtrix later. int gType: The way how the synaptic conductivity g will be defined. With GLOBALG we indicate that all conductance are of the same conductance value, which will be the value given in sPara. int delay: NO_DELAY means that there wil be no delays for synaptic signal propagation. int postSyn: Postsynaptic integration method, we are here using the standard model of an exponential decay of synaptic excitation. char *preName: Name of the pre-synaptic neuron population, here the Pop1 population. char *postName: Name of the post-synaptic neuron population, here also Pop1. double *sIni: A C-array of doubles containing initial values for the synaptic variables. double *sParam: A C-array of double precision that contains parameter values (common to all synapses of the population) double *psIni: A C-array of double precision numbers containing initial values for the post-synaptic model variables double *psPara: A C-array of double precision numbers containing parameters fo the post-snaptic model.Adding the addSynapsePopulation command to the model definition informs GeNN that there will be synapses between the named neuron populations, here between population Pop1 and itself. The detailed connectivity as defined by the variables g, we have still to define in the setup of our simulation. As always, the modelDefinition function ends on
At this point our model definition file tenHHRingModel.cc should look like this
Open the tenHHRingSimulation.cu file and update the file names of includes:
Now we need to add code to generate the desired ring connectivity.
After memory allocation and initialization gPop1self will contain only zeros. We then assign in the loop a non-zero conductivity of 0.01 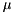 S to all synapses from neuron
S to all synapses from neuron i to i+1 (and 9 to 0 to close the ring).
After adjusting the GNUmakefile to read
we can build the model
and make it
After this there should be an exectable tenHHRingSimulation, which can be executed,
which should again result in
If we plot the content of columns one and two of tenHHexample.V.dat it looks very similar as in Tutorial 1
This is because the inhibitory synapses we created were all triggered at the same time so that they act during a post-synaptic spike which makes their effect all but invisible.
If we define different initial conditions for each of the ten neurons, i.e. add after initialize(),
then we observe different final values for each neuron,
and zooming in on the first 200 ms, the voltage of the first neuron now looks like this
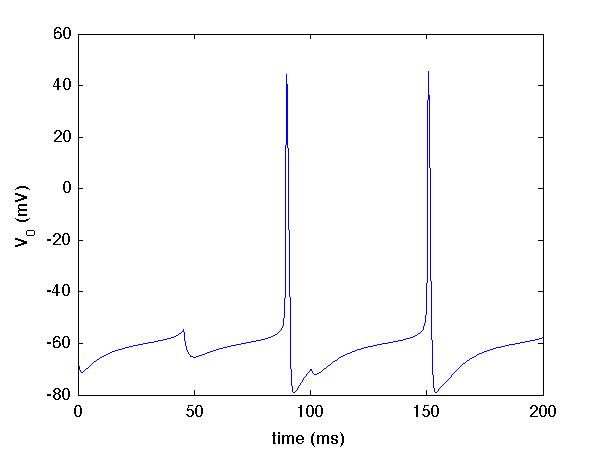
The complete codes for this tutorial are in userproject\tenHHRing_project.