 |
GeNN
2.2.3
GPU enhanced Neuronal Networks (GeNN)
|
 |
GeNN
2.2.3
GPU enhanced Neuronal Networks (GeNN)
|
There is a number of predefined models which can be chosen in the addNeuronGroup(...) function by their unique cardinal number, starting from 0. For convenience, C variables with readable names are predefined
The MAPNEURON type is a map based neuron model based on [4] but in the 1-dimensional map form used in [2] :
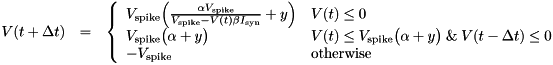
MAPNEURON type only works as intended for the single time step size of DT= 0.5.The MAPNEURON type has 2 variables:
V - the membrane potentialpreV - the membrane potential at the previous time stepand it has 4 parameters:
Vspike - determines the amplitude of spikes, typically -60mValpha - determines the shape of the iteration function, typically 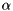 = 3
= 3y - "shift / excitation" parameter, also determines the iteration function,originally, y= -2.468beta - roughly speaking equivalent to the input resistance, i.e. it regulates the scale of the input into the neuron, typically 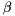 = 2.64
= 2.64  .
.MAPNEURON type needs two entries for V and Vpre and the parameter array needs four entries for Vspike, alpha, y and beta, in that order.Poisson neurons have constant membrane potential (Vrest) unless they are activated randomly to the Vspike value if (t- SpikeTime ) > trefract.
It has 3 variables:
V - Membrane potentialSeed - Seed for random number generationSpikeTime - Time at which the neuron spiked for the last timeand 4 parameters:
therate - Firing ratetrefract - Refractory periodVspike - Membrane potential at spike (mV)Vrest - Membrane potential at rest (mV)POISSONNEURON type needs three entries for V, Seed and SpikeTime and the parameter array needs four entries for therate, trefract, Vspike and Vrest, in that order.DT, i.e. the probability of firing is therate times DT: 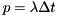 . This approximation is usually very good, especially for typical, quite small time steps and moderate firing rates. However, it is worth noting that the approximation becomes poor for very high firing rates and large time steps. An unrelated problem may occur with very low firing rates and small time steps. In that case it can occur that the firing probability is so small that the granularity of the 64 bit integer based random number generator begins to show. The effect manifests itself in that small changes in the firing rate do not seem to have an effect on the behaviour of the Poisson neurons because the numbers are so small that only if the random number is identical 0 a spike will be triggered.
. This approximation is usually very good, especially for typical, quite small time steps and moderate firing rates. However, it is worth noting that the approximation becomes poor for very high firing rates and large time steps. An unrelated problem may occur with very low firing rates and small time steps. In that case it can occur that the firing probability is so small that the granularity of the 64 bit integer based random number generator begins to show. The effect manifests itself in that small changes in the firing rate do not seem to have an effect on the behaviour of the Poisson neurons because the numbers are so small that only if the random number is identical 0 a spike will be triggered.seed variable. GeNN allocates memory for these seeds/states in the generated allocateMem() function. It is, however, currently the responsibility of the user to fill the array of seeds with actual random seeds. Not doing so carries the risk that all random number generators are seeded with the same seed ("0") and produce the same random numbers across neurons at each given time step. When using the GPU, seed also must be copied to the GPU after having been initialized.This conductance based model has been taken from [5] and can be described by the equations:
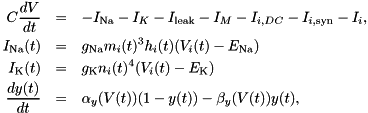
where 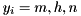 , and
, and
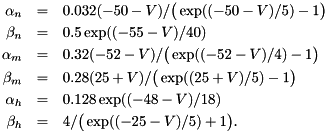
and typical parameters are 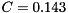 nF,
nF, 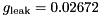
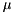 S,
S, 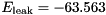 mV,
mV, 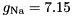
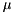 S,
S, 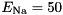 mV,
mV, 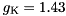
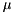 S,
S, 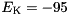 mV.
mV.
It has 4 variables:
V - membrane potential Em - probability for Na channel activation mh - probability for not Na channel blocking hn - probability for K channel activation nand 7 parameters:
gNa - Na conductance in 1/(mOhms * cm^2)ENa - Na equi potential in mVgK - K conductance in 1/(mOhms * cm^2)EK - K equi potential in mVgl - Leak conductance in 1/(mOhms * cm^2)El - Leak equi potential in mVCmem - Membrane capacity density in muF/cm^2DT= 0.1 ms, then the neurons are integrated with a linear Euler algorithm with lDT= 0.004 ms.Other variants of the same model are TRAUBMILES_ALTERNATIVE, TRAUBMILES_SAFE and TRAUBMILES_PSTEP. The former two are adressing the problem of singularities in the original Traub & Miles model [5]. At V= -52 mV, -25 mV, and -50 mV, the equations for 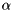 have denominators with value 0. Mathematically this is not a problem because the nominator is 0 as well and the left and right limits towards these singular points coincide. Numerically, however this does lead to nan (not-a-number) results through division by 0. The TRAUBMILES_ALTERNATIVE model adds SCALAR_MIN to the denominators at all times which typically is completely effect-free because it is truncated from teh mantissa, except when the denominator is very close to 0, in which case it avoids the singular value.
have denominators with value 0. Mathematically this is not a problem because the nominator is 0 as well and the left and right limits towards these singular points coincide. Numerically, however this does lead to nan (not-a-number) results through division by 0. The TRAUBMILES_ALTERNATIVE model adds SCALAR_MIN to the denominators at all times which typically is completely effect-free because it is truncated from teh mantissa, except when the denominator is very close to 0, in which case it avoids the singular value.
TRAUBMILES_SAFE takes a much more direct approach in which at the singular points, the correct value calculated offline with l'Hopital's rule in substituted. This is implemented with "if" statements.
Finally, the TRAUBMILES_PSTEP model allows users to control the number of internal loops, or sub-timesteps, that are used. This is enabled by making the number of time steps an explicit parameter of the model.
This is the Izhikevich model with fixed parameters [1]. It is usually described as
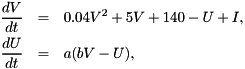
I is an external input current and the voltage V is reset to parameter c and U incremented by parameter d, whenever V >= 30 mV. This is paired with a particular integration procedure of two 0.5 ms Euler time steps for the V equation followed by one 1 ms time step of the U equation. Because of its popularity we provide this model in this form here event though due to the details of the usual implementation it is strictly speaking inconsistent with the displayed equations.
Variables are:
V - Membrane potentialU - Membrane recovery variableParameters are:
a - time scale of Ub - sensitivity of Uc - after-spike reset value of Vd - after-spike reset value of UThis is the same model as IZHIKEVICH (Izhikevich neurons with fixed parameters) IZHIKEVICH but parameters are defined as "variables" in order to allow users to provide individual values for each individual neuron instead of fixed values for all neurons across the population.
Accordingly, the model has the Variables:
V - Membrane potentialU - Membrane recovery variablea - time scale of Ub - sensitivity of Uc - after-spike reset value of Vd - after-spike reset value of Uand no parameters.
This model does not contain any update code and can be used to implement the equivalent of a SpikeGeneratorGroup in Brian or a SpikeSourceArray in PyNN.
In order to define a new neuron type for use in a GeNN application, it is necessary to populate an object of class neuronModel and append it to the global vector nModels. This can be done conveniently within the modelDefinition function just before the model is needed. The neuronModel class has several data members that make up the full description of the neuron model:
simCode of type string: This needs to be assigned a C++ (stl) string that contains the code for executing the integration of the model for one time step. Within this code string, variables need to be referred to by , where NAME is the name of the variable as defined in the vector varNames. The code may refer to the predefined primitives DT for the time step size and 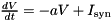 .
.thresholdConditionCode of type vector<string> (if applicable): Condition for true spike detection. supportCode of type string: This allows to define a code snippet that contains supporting code that will be utilized in the otehr code snippets. Typically, these are functions that are needed in the simCode or thresholdConditionCode. If possible, these should be defined as __host__ __device__ functions so that both GPU and CPU versions of GeNN code have an appropriate support code function available. The support code is protected with a namespace so that it is exclusively available for the neuronpopulation whose neurons define it. An example of a supportCode definition would be varNames of type vector<string>: This vector needs to be filled with the names of variables that make up the neuron state. The variables defined here as NAME can then be used in the syntax varTypes of type vector<string>: This vector needs to be filled with the variable type (e.g. "float", "double", etc) for the variables defined in varNames. Types and variables are matched to each other by position in the respective vectors, i.e. the 0th entry of varNames will have the type stored in the 0th entry of varTypes and so on. pNames of type vector<string>: This vector will contain the names of parameters relevant to the model. If defined as NAME here, they can then be referenced as a needed in the code example above.dpNames of type vector<string>: Names of "dependent
parameters". Dependent parameters are a mechanism for enhanced efficiency when running neuron models. If parameters with model-side meaning, such as time constants or conductances always appear in a certain combination in the model, then it is more efficient to pre-compute this combination and define it as a dependent parameter. This vector contains the names of such dependent parameters. 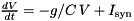 . Then one could define the code string and parameters as
. Then one could define the code string and parameters as dps of type dpclass*: The dependent parameter class, i.e. an implementation of dpclass which would return the value for dependent parameters when queried for them. E.g. in the example above it would return a when queried for dependent parameter 0 through dpclass::calculateDerivedParameter(). Examples how this is done can be found in the pre-defined classes, e.g. expDecayDp, pwSTDP, rulkovdp etc.extraGlobalNeuronKernelParameters of type vector<string>: On occasion, the neurons in a population share the same parameter. This could, for example, be a global reward signal. Such situations are implemented in GeNN with extraGlobalNeuronKernelParameters. This vector takes the names of such parameters. FOr each name, a variable will be created, with the name of the neuron population name appended, that can take a single value per population of the type defined in the extraGlobalNeuronKernelParameterTypes vector. This variable is available to all neurons in the population. It can also be used in synaptic code snippets; in this case it needs to be addressed with a _pre or _post postfix. For example, if teh pre-synaptic neuron population is of a neuron type that defines: s has a variable x and the synapse type s will only be used in conjunction with pre-synaptic neuron populations that do have the extraGlobalNeuronKernelParameter R. If the pre-synaptic population does not have the required variable/parameter, GeNN will fail when compiling the kernels.extraGlobalNeuronKernelParameterTypes of type vector<string>: These are the types of the extraGlobalNeuronKernelParameters. Types are matched to names by their position in the vector.Once the completed neuronModel object is appended to the nModels vector,
it can be used in network descriptions by referring to its cardinal number in the nModels vector. I.e., if the model is added as the 4th entry, it would be model "3" (counting starts at 0 in usual C convention). The information of the cardinal number of a new model can be obtained by referring to nModels.size() right before appending the new model, i.e. a typical use case would be.
. Then one can use the model as
In earlier versions of GeNN External input to a neuron group could be activated by calling the activateDirectInput function. This was now removed in favour of defining a new neuron model where the direct input can be a parameter (constant over time and homogeneous across the population), a variable (changing in time and non-homogeneous across the population), or an extraGlobalNeuronKernelParameter (changing in time but homogeneous across the population). How this can be done is illustrated for example in the Izh_sparse example project.