 |
GeNN
2.2.3
GPU enhanced Neuronal Networks (GeNN)
|
 |
GeNN
2.2.3
GPU enhanced Neuronal Networks (GeNN)
|
A synapse model is a weightUpdateModel object which consists of variables, parameters, and string objects which will be substituted in the code. The three strings that will be substituted in the code for synapse update are:
simCode: Simulation code that is used when a true spike is detected. The update is performed only once, one time step after threshold condition detection, which is defined by thresholdConditionCode in the neuron model of the corresponding presynaptic neuron population. evntThreshold: Code that defines the condition for a synaptic event. This typically involves the pre-synaptic variables, e.g. the membrane potential: simCodeEvnt is executed. For an example, see NGRADSYNAPSE (Graded Synapse).simCodeEvnt: Simulation code that is used for spike like events, where updates are done for all instances in which the event condition defined by evntThreshold is met. evntThreshold is also be provided as a code string. For an example, see NGRADSYNAPSE (Graded Synapse).simLearnPost: Simulation code which is used in the learnSynapsesPost kernel/function, which performs updates to synapses that are triggered by post-synaptic spikes. This is rather unusual other than in learning rules like e.g. STDP. For an example that uses simLearnPost, see LEARN1SYNAPSE (Learning Synapse with a Primitive Piece-wise Linear Rule).synapseDynamics: Simulation code that applies for every time step, i.e.is unlike the others not gated with a condition. This can be used where synapses have internal variables and dynamics that are described in continuous time, e.g. by ODEs. Usng this mechnanism is typically computationally veruy costly because of the large number of synapses in a typical network.extraGlobalSynapseKernelParameters of type vector<string>: On occasion, the synapses in a synapse population share a global parameter. This could, for example, be a global reward signal. This is supported in GeNN with extraGlobalSynapseKernelParameters. The user defines the names of such parameters and pushes them into this vector. GeNN creates variables of this name, with the name of the synapse population appended, that can take a single value per population of the type defined in the extraGlobalSynapseKernelParameterTypes vector. This variable is then available to all synapses in the population. extraGlobalSynapseKernelParameters is necessary as they are communicated as kernel parameters.extraGlobalSynapseKernelParameterTypes of type vector<string>: These are the types of the extraGlobalSynapseKernelParameters. Types are matched to names by their position in the vector.All code snippets can be used to manipulate any synapse variable and so implement both synaptic dynamics and learning processes.
Currently 3 predefined synapse models are available:
These are defined in lib.include/utils.h. The MBody_userdef example also includes a modified version of these models as user-defined models.
If this model is selected, no learning rule is applied to the synapse and for each pre-synaptic spikes the synaptic conductances are simply added to the postsynaptic input variable. The model has 1 variable:
g - conductance of scalar typeand no other parameters.
simCode is:
In a graded synapse, the conductance is updated gradually with the rule:
![\[ gSyn= g * tanh((V - E_{pre}) / V_{slope} \]](form_20.png)
whenever the membrane potential 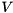 is larger than the threshold
is larger than the threshold 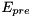 . The model has 1 variable:
. The model has 1 variable:
g: conductance of scalar typeThe parameters are:
Epre: Presynaptic threshold potentialVslope: Activation slope of graded releasesimCodeEvnt is:
evntThreshold is:
_pre in synapse related code such as an evntThreshold. Users can also access post-synaptic neuron variables using the suffix _post.This is a simple STDP rule including a time delay for the finite transmission speed of the synapse, defined as a piecewise function:
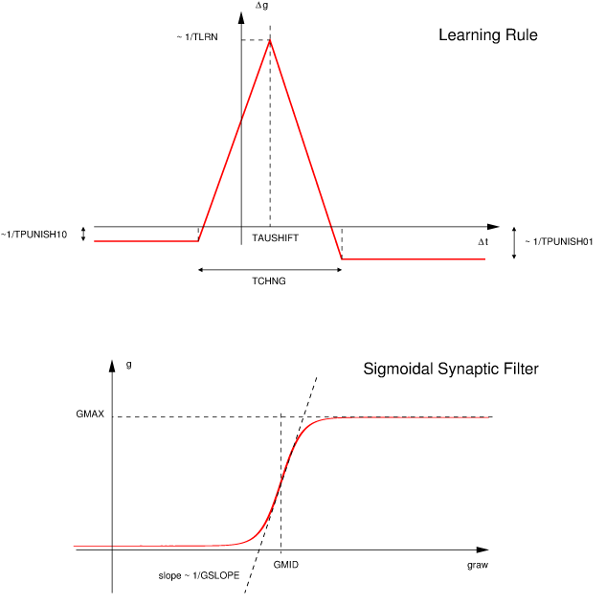
The STDP curve is applied to the raw synaptic conductance gRaw, which is then filtered through the sugmoidal filter displayed above to obtain the value of g.
gRaw (and hence in g).gRaw that is subject to the STDP rule. The resulting synaptic conductance is a sigmoid filter of gRaw. This implies that g is initialised but not gRaw, the synapse will revert to the value that corresponds to gRaw.An example how to use this synapse correctly is given in map_classol.cc (MBody1 userproject):
g fully to 0, as this leads to gRaw= -infinity and this is not support. I.e., 'g' needs to be some nominal value > 0 (but can be extremely small so that it acts like it's 0).The model has 2 variables:
g: conductance of scalar typegRaw: raw conductance of scalar typeParameters are (compare to the figure above):
Epre: Presynaptic threshold potentialtLrn: Time scale of learning changestChng: Width of learning windowtDecay: Time scale of synaptic strength decaytPunish10: Time window of suppression in response to 1/0tPunish01: Time window of suppression in response to 0/1gMax: Maximal conductance achievablegMid: Midpoint of sigmoid g filter curvegSlope: Slope of sigmoid g filter curvetauShift: Shift of learning curvegSyn0: Value of syn conductance g decays toFor more details about these built-in synapse models, see [3].
If users want to define their own models, they can add a new weightUpdateModel that includes the variables, parameters, and update codes as desired, and then push this object in the weightUpdateModels vector. The model can be used by referring to its index in the weightUpdateModels vector when adding a new population by with a call to addSynapsePopulation.
The available options work as follows:
INDIVIDUALG: When this option is chosen in the addSynapsePopulation command, GeNN reserves an array of size n_pre x n_post float for individual conductance values for each combination of pre and postsynaptic neuron. The actual values of the conductances are passed at runtime from the user side code, using the pushXXXXXToDevice function, where XXXX is the name of the synapse population.GLOBALG: When this option is chosen, the value of the variables of the synapse model (including its conductance) is taken to be the initial value provided for the synapse model's variables. This option can only be sensibly combined with connectivity type ALLTOALL.INDIVIDUALID: When this option is chosen, GeNN expects to use the same maximal conductance for all existing synaptic connections but which synapses exist will be defined at runtime from the user side code, provided as a binary array (see Insect olfaction model).Available options are DENSE and SPARSE. Various tools are provided under userproject/tools for creating different connectivity schemes.
Different strategies are used by GeNN for different combinations of connecticity types and Conductance definition methods, as explained in the table below:
| ALLTOALL | DENSE | SPARSE | |
|---|---|---|---|
| GLOBALG | Fixed values for all synapse members | Fixed values for all synapse members | Fixed values for some synapse members (using sparse indexing) |
| INDIVIDUALG | Variable values for all synapse members | Variable values for all synapse members | Variable values for some synapse members |
| INDIVIDUALID | Fixed values for some synapse members (using a binary array). Technically possible but not meaningful. | Fixed values for some synapse members (using a binary array) | N/A |
If INDIVIDUALG is used with ALLTOALL or DENSE connectivity (these are equivalent in this case), synapse variables are stored in an array of size npre * npost .
If the connectivity is of SPARSE type, connectivity indices are stored in a struct named SparseProjection in order to minimize the memory requirements. The struct SparseProjection contains the following members: 1: unsigned int connN: number of connections in the population. This value is needed for allocation of arrays. The indices that correspond to these values are defined in a pre-to-post basis by the following arrays: 2: unsigned int ind, of size connN: Indices of corresponding postsynaptic neurons concatenated for each presynaptic neuron. 3: unsigned int *indInG, of size model.neuronN[model.synapseSource[synInd]]+1: This array defines from which index in the synapse variable array the indices in ind would correspond to the presynaptic neuron that corresponds to the index of the indInG array, with the number of connections being the size of ind. More specifically, indIng[n+1]-indIng[n] would give the number of postsynaptic connections for neuron n.
For example, consider a network of two presynaptic neurons connected to three postsynaptic neurons: 0th presynaptic neuron connected to 1st and 2nd postsynaptic neurons, the 1st presynaptic neuron connected to 0th and 2nd neurons. The struct SparseProjection should have these members, with indexing from 0:
ConnN = 4
ind= [1 2 0 2]
indIng= [0 2 4]
A synapse variable of a sparsely connected synapse will be kept in an array using this conductance for indexing. For example, a variable caled g will be kept in an array such as: g=[g_Pre0-Post1 g_pre0-post2 g_pre1-post0 g_pre1-post2] If there are no connections for a presynaptic neuron, then g[indIng[n]]=gp[indIng[n]+1].
See tools/gen_syns_sparse_IzhModel used in Izh_sparse project to see a working example.
The postSynModel defines how synaptic activation translates into an input current (or other input term for models that are not current based). It also can contain equations defining dynamics that are applied to the (summed) synaptic activation, e.g. an exponential decay over time.
A postSynModel object consists of variables, parameters, derived parameters and two strings that define the code for current generation and continuous dynamics.
postSynDecay: This code defines the continuous time dynamics of the summed presynaptic inputs at the postsynaptic neuron. This usually consists of some kind of decay function.postSyntoCurrent: This code defines how the synaptic inputs lead to an input input current (Isyn) to the postsynaptic neuron.There are currently 2 built-in postsynaptic integration methods:
EXPDECAY: Exponential decay. Decay time constant and reversal potential parameters are needed for this postsynaptic mechanism.
This model has no variables and two parameters:
tau : Decay time constantE : Reversal potentialtau is used by the derived parameter expdecay which returns expf(-dt/tau).
IZHIKEVICH_PS: Empty postsynaptic rule to be used with Izhikevich neurons.