 |
GeNN
4.9.0
GPU enhanced Neuronal Networks (GeNN)
|
 |
GeNN
4.9.0
GPU enhanced Neuronal Networks (GeNN)
|
Hodgkin-Huxley neurons with Traub & Miles algorithm. More...
#include <neuronModels.h>
Public Types | |
| typedef Snippet::ValueBase< 7 > | ParamValues |
| typedef Models::VarInitContainerBase< 4 > | VarValues |
| typedef Models::VarInitContainerBase< 0 > | PreVarValues |
| typedef Models::VarInitContainerBase< 0 > | PostVarValues |
 Public Types inherited from Models::Base Public Types inherited from Models::Base | |
| typedef std::vector< Var > | VarVec |
| typedef std::vector< VarRef > | VarRefVec |
| typedef std::vector< EGPRef > | EGPRefVec |
 Public Types inherited from Snippet::Base Public Types inherited from Snippet::Base | |
| typedef std::vector< std::string > | StringVec |
| typedef std::vector< EGP > | EGPVec |
| typedef std::vector< ParamVal > | ParamValVec |
| typedef std::vector< DerivedParam > | DerivedParamVec |
Public Member Functions | |
| virtual std::string | getSimCode () const override |
| Gets the code that defines the execution of one timestep of integration of the neuron model. More... | |
| virtual std::string | getThresholdConditionCode () const override |
| Gets code which defines the condition for a true spike in the described neuron model. More... | |
| virtual StringVec | getParamNames () const override |
| Gets names of of (independent) model parameters. More... | |
| virtual VarVec | getVars () const override |
| Gets names and types (as strings) of model variables. More... | |
 Public Member Functions inherited from NeuronModels::Base Public Member Functions inherited from NeuronModels::Base | |
| virtual std::string | getResetCode () const |
| Gets code that defines the reset action taken after a spike occurred. This can be empty. More... | |
| virtual std::string | getSupportCode () const |
| Gets support code to be made available within the neuron kernel/funcion. More... | |
| virtual Models::Base::ParamValVec | getAdditionalInputVars () const |
| virtual bool | isAutoRefractoryRequired () const |
| Does this model require auto-refractory logic? More... | |
| boost::uuids::detail::sha1::digest_type | getHashDigest () const |
| Update hash from model. More... | |
| void | validate () const |
| Validate names of parameters etc. More... | |
 Public Member Functions inherited from Models::Base Public Member Functions inherited from Models::Base | |
| size_t | getVarIndex (const std::string &varName) const |
| Find the index of a named variable. More... | |
 Public Member Functions inherited from Snippet::Base Public Member Functions inherited from Snippet::Base | |
| virtual | ~Base () |
| virtual DerivedParamVec | getDerivedParams () const |
| virtual EGPVec | getExtraGlobalParams () const |
| size_t | getExtraGlobalParamIndex (const std::string ¶mName) const |
| Find the index of a named extra global parameter. More... | |
Static Public Member Functions | |
| static const NeuronModels::TraubMiles * | getInstance () |
Additional Inherited Members | |
 Protected Member Functions inherited from Models::Base Protected Member Functions inherited from Models::Base | |
| void | updateHash (boost::uuids::detail::sha1 &hash) const |
| void | validate () const |
| Validate names of parameters etc. More... | |
 Protected Member Functions inherited from Snippet::Base Protected Member Functions inherited from Snippet::Base | |
| void | updateHash (boost::uuids::detail::sha1 &hash) const |
| void | validate () const |
| Validate names of parameters etc. More... | |
 Static Protected Member Functions inherited from Snippet::Base Static Protected Member Functions inherited from Snippet::Base | |
| template<typename T > | |
| static size_t | getNamedVecIndex (const std::string &name, const std::vector< T > &vec) |
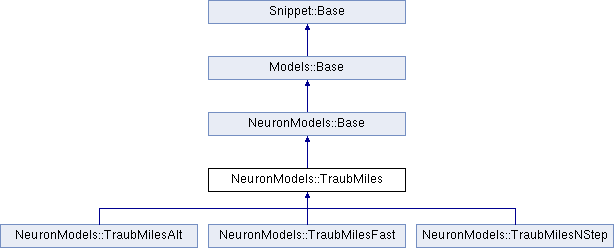
Hodgkin-Huxley neurons with Traub & Miles algorithm.
This conductance based model has been taken from [9] and can be described by the equations:
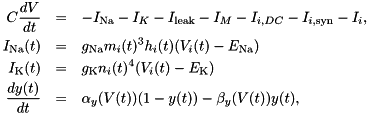
where 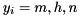 , and
, and
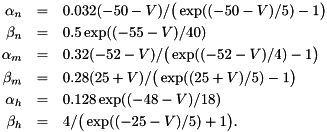
and typical parameters are 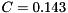 nF,
nF, 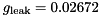
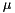 S,
S, 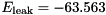 mV,
mV, 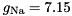
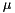 S,
S, 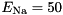 mV,
mV, 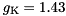
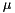 S,
S, 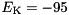 mV.
mV.
It has 4 variables:
V - membrane potential Em - probability for Na channel activation mh - probability for not Na channel blocking hn - probability for K channel activation nand 7 parameters:
gNa - Na conductance in 1/(mOhms * cm^2)ENa - Na equi potential in mVgK - K conductance in 1/(mOhms * cm^2)EK - K equi potential in mVgl - Leak conductance in 1/(mOhms * cm^2)El - Leak equi potential in mVC - Membrane capacity density in muF/cm^2DT= 0.1 ms, then the neurons are integrated with a linear Euler algorithm with lDT= 0.004 ms. This variant uses IF statements to check for a value at which a singularity would be hit. If so, value calculated by L'Hospital rule is used. | typedef Snippet::ValueBase< 7 > NeuronModels::TraubMiles::ParamValues |
|
inlinestatic |
|
inlineoverridevirtual |
Gets names of of (independent) model parameters.
Reimplemented from Snippet::Base.
Reimplemented in NeuronModels::TraubMilesNStep.
|
inlineoverridevirtual |
Gets the code that defines the execution of one timestep of integration of the neuron model.
The code will refer to for the value of the variable with name "NN". It needs to refer to the predefined variable "ISYN", i.e. contain , if it is to receive input.
Reimplemented from NeuronModels::Base.
Reimplemented in NeuronModels::TraubMilesNStep, NeuronModels::TraubMilesAlt, and NeuronModels::TraubMilesFast.
|
inlineoverridevirtual |
Gets code which defines the condition for a true spike in the described neuron model.
This evaluates to a bool (e.g. "V > 20").
Reimplemented from NeuronModels::Base.
|
inlineoverridevirtual |
Gets names and types (as strings) of model variables.
Reimplemented from Models::Base.