 |
GeNN
3.3.0
GPU enhanced Neuronal Networks (GeNN)
|
 |
GeNN
3.3.0
GPU enhanced Neuronal Networks (GeNN)
|
There is a number of predefined models which can be used with the NNmodel::addNeuronGroup function:
In order to define a new neuron type for use in a GeNN application, it is necessary to define a new class derived from NeuronModels::Base. For convenience the methods this class should implement can be implemented using macros:
DT for the time step size and NAME here, they can then be referenced as $(NAME) in the code string. The length of this list should match the NUM_PARAM specified in DECLARE_MODEL. Parameters are assumed to be always of type double.NAME can then be used in the syntax $(NAME) in the code string.For example, using these macros, we can define a leaky integrator 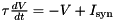 solved using Euler's method:
solved using Euler's method:
Additionally "dependent parameters" can be defined. Dependent parameters are a mechanism for enhanced efficiency when running neuron models. If parameters with model-side meaning, such as time constants or conductances always appear in a certain combination in the model, then it is more efficient to pre-compute this combination and define it as a dependent parameter.
For example, because the equation defining the previous leaky integrator example has an algebraic solution, it can be more accurately solved as follows - using a derived parameter to calculate 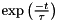 :
:
GeNN provides several additional features that might be useful when defining more complex neuron models.
Support code enables a code block to be defined that contains supporting code that will be utilized in multiple pieces of user code. Typically, these are functions that are needed in the sim code or threshold condition code. If possible, these should be defined as __host__ __device__ functions so that both GPU and CPU versions of GeNN code have an appropriate support code function available. The support code is protected with a namespace so that it is exclusively available for the neuron population whose neurons define it. Support code is added to a model using the SET_SUPPORT_CODE() macro, for example:
Extra global parameters are parameters common to all neurons in the population. However, unlike the standard neuron parameters, they can be varied at runtime meaning they could, for example, be used to provide a global reward signal. These parameters are defined by using the SET_EXTRA_GLOBAL_PARAMS() macro to specify a list of variable names and type strings (like the SET_VARS() macro). For example:
These variables are available to all neurons in the population. They can also be used in synaptic code snippets; in this case it need to be addressed with a _pre or _post postfix.
For example, if the model with the "R" parameter was used for the pre-synaptic neuron population, the weight update model of a synapse population could have simulation code like:
where we have assumed that the weight update model has a variable x and our synapse type will only be used in conjunction with pre-synaptic neuron populations that do have the extra global parameter R. If the pre-synaptic population does not have the required variable/parameter, GeNN will fail when compiling the kernels.
Normally, neuron models receive the linear sum of the inputs coming from all of their synaptic inputs through the $(inSyn) variable. However neuron models can define additional input variables - allowing input from different synaptic inputs to be combined non-linearly. For example, if we wanted our leaky integrator to operate on the the product of two input currents, it could be defined as follows:
Where the SET_ADDITIONAL_INPUT_VARS() macro defines the name, type and its initial value before postsynaptic inputs are applyed (see section Postsynaptic integration methods for more details).
Many neuron models have probabilistic terms, for example a source of noise or a probabilistic spiking mechanism. In GeNN this can be implemented by using the following functions in blocks of model code:
$(gennrand_uniform) returns a number drawn uniformly from the interval ![$[0.0, 1.0]$](../../form_2.png)
$(gennrand_normal) returns a number drawn from a normal distribution with a mean of 0 and a standard deviation of 1.$(gennrand_exponential) returns a number drawn from an exponential distribution with 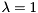 .
.$(gennrand_log_normal, MEAN, STDDEV) returns a number drawn from a log-normal distribution with the specified mean and standard deviation.$(gennrand_gamma, ALPHA) returns a number drawn from a gamma distribution with the specified shape.Once defined in this way, new neuron models classes, can be used in network descriptions by referring to their type e.g.